
- SUPERPRO DESIGNER SOFTWARE .DLL
- SUPERPRO DESIGNER SOFTWARE INSTALL
- SUPERPRO DESIGNER SOFTWARE PATCH
- SUPERPRO DESIGNER SOFTWARE FULL
Please note that if you don't have Administrative Privileges, the copying of the files may fail.

It will assume that the location is the default suggested by the original installation script ("C:\Program Files (x86)\Intelligen\SuperPro Designer\v") but if you have chosen a different location, you must provide it (there will be a prompt for that).
SUPERPRO DESIGNER SOFTWARE .DLL
dll files) and copy them into the folder where the software is currently installed. As it runs, it will extract all its contained files (one. Right click on it and select 'Run As Administrator'. The file you downloaded is self-expanding. It does NOT contain the entire contents of the installation CD for this edition of the software. The file contains in a compressed format, only the runtime files that have changed in this release. Save the file on a local drive of your PC that is NOT a mapped drive.
SUPERPRO DESIGNER SOFTWARE PATCH
If You Downloaded a Patch Installation Packet. You should keep the contents of this folder in case you need to access an individual file from the installation set that may be accidentally deleted or corrupt (e.g. If you wish to create an installation CD, you can copy all the files under the "SPDv12 Installer" on a blanc CD.
SUPERPRO DESIGNER SOFTWARE INSTALL
Simply follow the instructions to install the software.

After all the files are extracted into the "SPDv12 Installer" subfolder, the installation scipt will be automatically started. It will expand all its contained files into a subfolder named "SPDv12 Installer" but you can change it if desired (there will be a prompt for that). Since the expanded file structure is quite deep, if you started from a location that is more than 2-level deep, it is possible that Windows may not be able to access all the installation files in order to copy them in the target location(s). The file contains in a compressed format, the entire contents of the installation CD for this edition of the software. Save the file on a local physical drive of your PC that is NOT a mapped drive and in a folder that is NOT more than 2-levels deep.
SUPERPRO DESIGNER SOFTWARE FULL
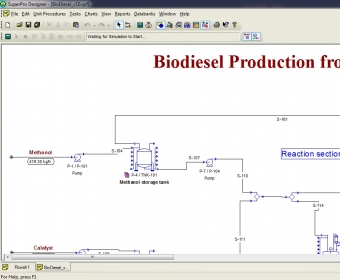
If You Downloaded a Full Installation Packet in a Single File. Major Version 13, Build 01, SBN 1065 (Rel. publication), you must acknowledge the authors of this publication in your work.īY DOWNLOADING OR USING THE SUPERPRO DESIGNER ® FLOWSHEET FILES, AND/OR CLICKING THE “I AGREE” BUTTON, YOU ARE INDICATING YOUR ACCEPTANCE OF THE TERMS AND CONDITIONS HEREIN.More info for administrators can be found in the Sys Admin page. If you use the SuperPro Designer ® Flowsheet Files in any means as an aid to your work (e.g. The McDonald/Nandi Lab grants you, and you hereby accept, a non-exclusive, non-transferable, royalty-free perpetual agreement to download and use the version noted above of the SuperPro Designer ® Flowsheet Files noted above.Ģ. IMPORTANT – READ CAREFULLY: THIS END-USER AGREEMENT IS AN AGREEMENT BETWEEN YOU AND THE MCDONALD/NANDI LAB.ġ. SuperPro Designer ® Flowsheet Files: Plant-Based Production of Griffithsin Techno-Economic Analysis Upstream and Downstream Processing SuperPro Designer ® Flowsheet Files (.spf Files), Version: v.1 STEP 2: DOWNLOAD SUPERPRO DESIGNER ® FLOWSHEET FILES For more information about the software requirements or specifications, please contact Intelligen, Inc. If you do not own a license to the software, you may download a free trial version from Intelligen, Inc.’s website. If you already own a license to the software, please GO DIRECTLY TO STEP 2. A simulation in bioprocess technology was carried out in order to evaluate the accessibility and adequacy of the simulation software SuperProDesigner used.
In order to run the models, SuperPro Designer ® must be installed on your computer. Please follow the instructions below to download the models. Frontiers in Bioengineering and Biotechnology, 6, 102 Technoeconomic Modeling of Plant-Based Griffithsin Manufacturing. Thank you for interest in our publication,


 0 kommentar(er)
0 kommentar(er)
